新闻动态 News
查看更多 >>
-
2021-07-052021年6月30日,北京望石智慧科技有限公司(以下简称“望石智慧”)与北京泰德制药股份有限公司(以下简称“泰德制药”)宣布双方达成进一步合作。 首次合作中,双方针对某国际领先肿瘤靶点进行小分子药物的共同开发。望石智慧利用复合的临床前研发团队、AI分子生成平台和超高通量筛选平台,在短时间内获得了数个有专利空间、活性达成药性级别的全新化学骨架,极大程度提高了早期研发速度,且分子合成数量少,总试验成本低。 双方基于对进展的认可决定进行更深层次的合作,包括对已有项目加速开发,同时进行更深度的商业模式探索和利益绑定。 望石智慧团队的复合型、对药物的深度理解和体系化的AI平台是双方早期合作中展示出的实力,结合泰德在创新药领域的大力投入、对于靶点的布局和高质量的研发平台,双方取得的成果较为喜人。我们也愿意跟望石智慧更深度的合作,更好的整合双方的优势和资源,开启长久的伙伴关系。——泰德制药研发副总裁赵焰平 泰德的研发中心每年在创新药的研发投入大量资金和精力。前期合...
-
2021-04-112021年4月12日,人工智能技术驱动创新药研发的平台型科技公司——望石智慧(StoneWise)宣布完成了B+轮和B轮融资,融资总额一亿美元。B+轮融资由大湾区共同家园发展基金和光速中国共同领投,B轮融资由君联资本领投。新进股东鼎晖VGC(创新与成长基金)和钟鼎资本,老股东高瓴创投、SIG海纳亚洲、长岭资本、线性资本等共同参与。此次融资中,望石智慧A轮、A+轮股东全线跟投,其中高瓴创投进行了超额跟投。跃为资本担任本次融资的独家财务顾问。 融资将主要用于进一步吸引全球顶尖人才,巩固望石AI平台的产业化落地成果,加快望石AI技术创新的迭代速度,提升赋能效率。 凭借已被验证的商业模式、夯实的技术根基、优秀的团队,望石智慧在过去一年内连续完成四轮融资,投资方同时涵盖医药和AI两个领域的头部机构,在业内较为少见。此次融资标志着望石智慧打造的“分子设计平台+知识图谱”双引擎,在数据飞轮的驱动下加速了“AI平台赋能小分子药物研发”产业化落地的成功。 开发周期长、风险大、投资高、失败率...
-
2020-09-169月8日,第八届海外人才创业大会(OTEC 2020)全球创业赛抗疫创业赛道决赛在北京落幕,望石智慧凭借在抗击疫情中扎实的新冠药物筛选工作、领先的医药分子研发技术、攻坚克难的科技创新精神,从众多优秀参赛项目中脱颖而出,荣获“抗疫创新先锋”奖。 (OTEC2020抗疫创业大赛颁奖现场) 2020年,是全球共抗疫情的一年,更是人类需要共同应对“后疫情时代”危与机并存的一年。为鼓励在疫情中做出突出贡献的企业和人才,由北京市朝阳区打造的服务海外人才创新创业的OTEC2020全球创业赛特别推出了抗疫创业赛道。在本场赛道的比拼中,创业者们充分展示了抗疫项目的社会价值,彰显了新一代年轻企业家的风采,传递了抗疫创新能量——创新将引领“后疫情时代”发展! (CEO助理兼药化总监张宏波博士代表望石智慧参与决赛) 面对突如其来的疫情,望石智慧在今年1月下旬时紧急成立了抗击新冠项目组,在一周内完成了2019-nCOV 靶向蛋白RDRP的构象预测,并利用知识图谱快速提取超过140...
-
2020-09-099月9日,北京望石智慧科技有限公司(以下简称“望石智慧”)宣布,公司近期已完成数百万美金的A+轮融资,投资方为高瓴创投(GL Ventures),现有投资股东线性资本超额投资,明希资本担任财务顾问。本轮融资资金将用于持续优化AI技术与药物发现技术的嫁接与创新、搭建世界领先的智能化药物研发平台和经验丰富的复合型人才团队,同时进一步推进海内外战略合作与业务拓展。 望石智慧成立以来已完成三轮融资,分别是2018年底由SIG投资的天使轮,2020年3月由长岭资本和线性资本投资的A轮,以及本次高瓴创投投资的A+轮,标志着望石智慧致力于打造的基于AI技术的“软件+里程碑”一体化小分子创新药设计解决方案在市场上可谓再度得到认可。 全球创新药临床前研发每年投入达500亿美元以上,且仍呈增长趋势,从临床I期不同药物或疗法试验分布来看,超过65%仍为小分子药物,而小分子的临床前研发本质是一个提出假设与试错的过程,这意味着其潜在压缩空间巨大。全球和中国本土创新药企正在积极尝试拥抱并利用AI技术提高小...
学术进展 Academic Progress
查看更多 >>
-
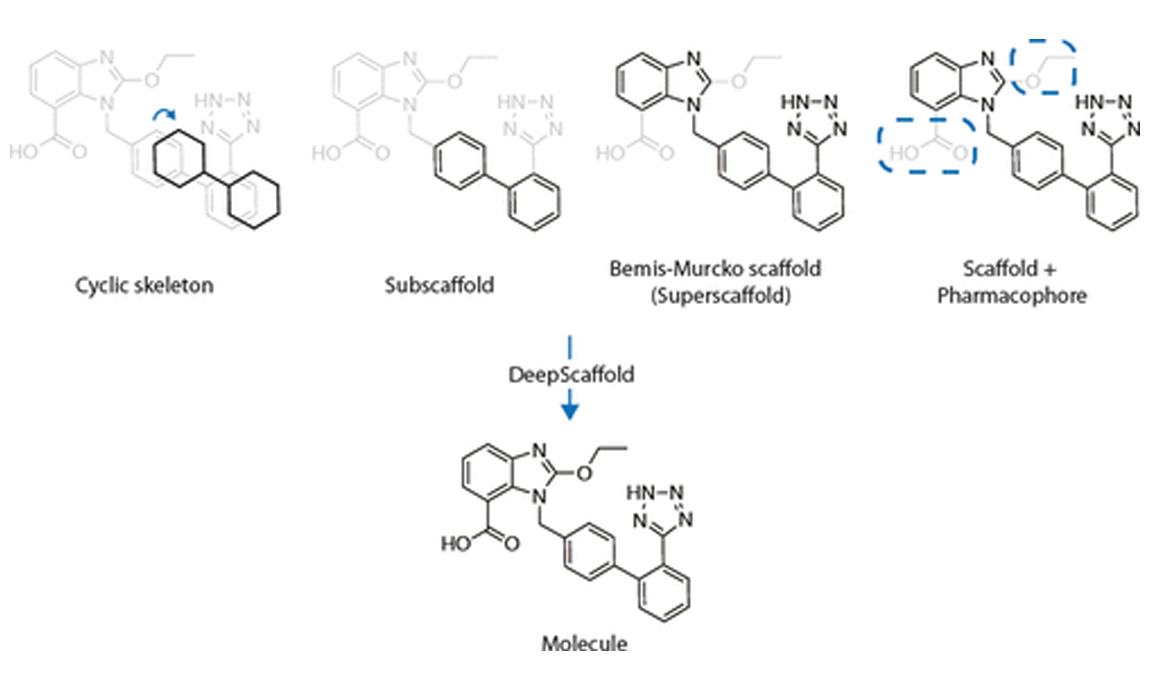
2022-09-15Yibo Li, Jianxing Hu, Yanxing Wang, Jielong Zhou, Liangren Zhang, and Zhenming Liu J Chem Inf Model 2020 Jan 27;60(1):77-91. doi: 10.1021/acs.jcim.9b00727. Epub 2019 Dec 20. Abstract The ultimate goal of drug design is to find novel compounds with desirable pharmacological properties. Designing molecules retaining particular scaffolds as their core structures is an efficient way to obtain potential drug candidates. We propose a scaffold-based molecular generative model for drug discovery, which performs molecule generation based on a wide spectrum of scaffold definitions, including Bemis-Murcko scaffolds, cyclic skeletons, and scaffolds with specifications on side-chain properties. The model can generalize the learned chemical rules of adding atoms and bonds to a given scaffold. The generated compounds were evaluated by molecular docking in DRD2 targets, and...
-
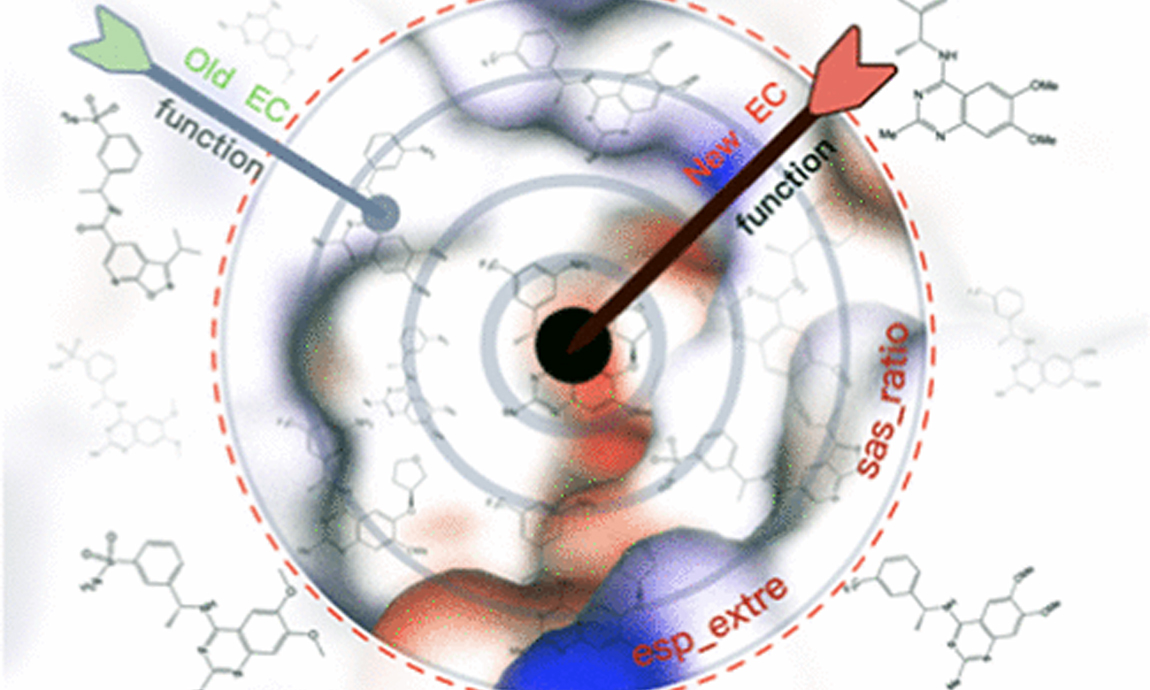
2022-09-15Liming Zhao, Mengchen Pu, Huting Wang, Xiangyu Ma, and Yingsheng J. Zhang J Chem Inf Model. 2022 Sep 7. doi: 10.1021/acs.jcim.2c00616. Abstract In recent years, machine learning (ML) models have been found to quickly predict various molecular properties with accuracy comparable to high-level quantum chemistry methods. One such example is the calculation of electrostatic potential (ESP). Different ESP prediction ML models were proposed to generate surface molecular charge distribution. Electrostatic complementarity (EC) can apply ESP data to quantify the complementarity between a ligand and its binding pocket, leading to the potential to increase the efficiency of drug design. However, there is not much research discussing EC score functions and their applicability domain. We propose a new EC score function modified from the one originally develop...
-
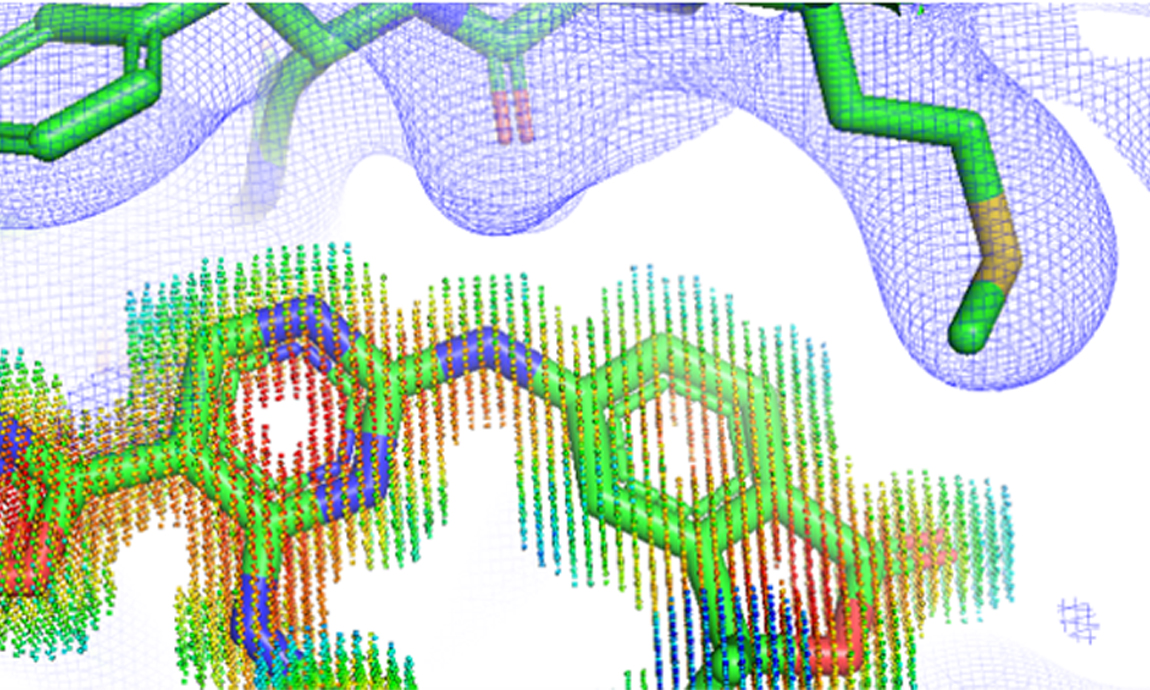
2022-09-15Sci Rep. 2022 Sep 6;12(1):15100. doi: 10.1038/s41598-022-19363-6. Lvwei Wang, Rong Bai, Xiaoxuan Shi, Wei Zhang, Yinuo Cui, Xiaoman Wang, Cheng Wang, Haoyu Chang, Yingsheng Zhang, Jielong Zhou, Wei Peng, Wenbiao Zhou & Bo Huang Abstract We report for the first time the use of experimental electron density (ED) as training data for the generation of drug-like three-dimensional molecules based on the structure of a target protein pocket. Similar to a structural biologist building molecules based on their ED, our model functions with two main components: a generative adversarial network (GAN) to generate the ligand ED in the input pocket and an ED interpretation module for molecule generation. The model was tested on three targets: a kinase (hematopoietic progenitor kinase 1), protease (SARS-CoV-2 main protease), and nuclear receptor (vi...
-
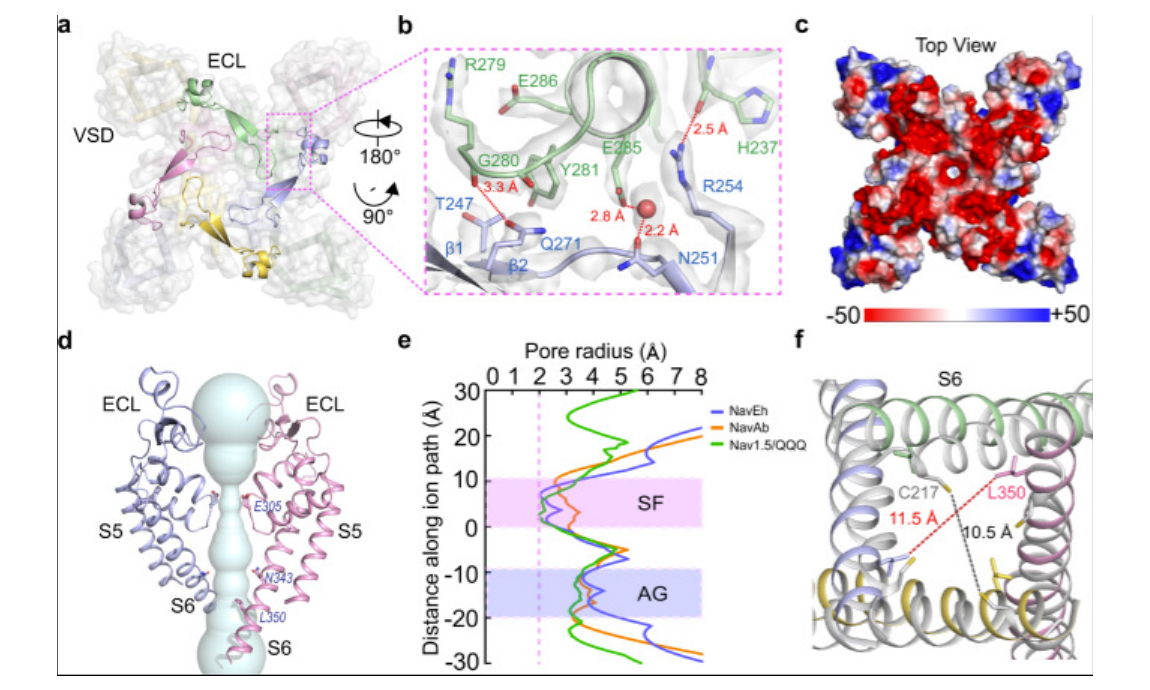
2022-06-06Jiangtao Zhang, Yiqiang Shi, Junping Fan, Huiwen Chen, Zhanyi Xia, Bo Huang, Juquan Jiang, Jianke Gong, Zhuo Huang, Daohua Jiang Abstract: Voltage-gated sodium (NaV) channels initiate action potentials. Fast inactivation of NaV channels, mediated by an Ile-Phe-Met motif, is crucial for preventing hyperexcitability and regulating firing frequency. Here we present cryo-electron microscopy structure of NaVEh from the coccolithophore Emiliania huxleyi, which reveals an unexpected molecular gating mechanism for NaV channel fast inactivation independent of the Ile-Phe-Met motif. An N-terminal helix of NaVEh plugs into the open activation gate and blocks it. The binding pose of the helix is stabilized by multiple electrostatic interactions. Deletion of the helix or mutations blocking the electrostatic interactions completely abolish...
-
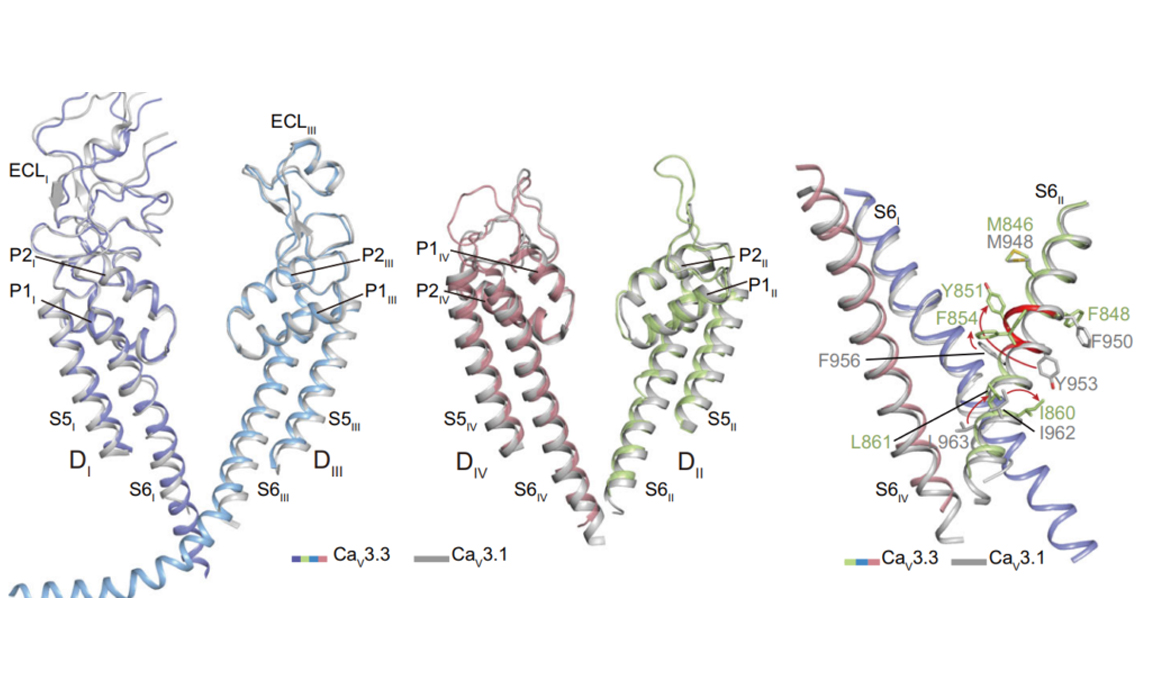
2022-04-27Abstract: The low-voltage activated T-type calcium channels regulate cellular excitability and oscillatory behavior of resting membrane potential which trigger many physiological events and have been implicated with many diseases. Here, we determine structures of the human T-type CaV3.3 channel, in the absence and presence of antihypertensive drug mibefradil, antispasmodic drug otilonium bromide and antipsychotic drug pimozide. CaV3.3 contains a long bended S6 helix from domain III, with a positive charged region protruding into the cytosol, which is critical for T-type CaV channel activation at low voltage. The drug-bound structures clearly illustrate how these structurally different compounds bind to the same central cavity inside the CaV3.3 channel, but are mediated by significantly distinct interactions between drugs and their surrounding residues. Pho...
-
2022-04-27Abstract: We report for the first time the use of experimental electron density (ED) in the Protein Data Bank for modeling of noncovalent interactions (NCIs) for protein–ligand complexes. Our methodology is based on reduced electron density gradient (RDG) theory describing intermolecular NCIs by ED and its first derivative. We established a database named Experimental NCI Database (ExptNCI; http://ncidatabase.stonewise.cn/#/nci) containing ED saddle points, indicating ∼200,000 NCIs from over 12,000 protein–ligand complexes. We also demonstrated the usage of the database in the case of depicting amide−π interactions in protein–ligand binding systems. In summary, the database provides details on experimentally observed NCIs for protein–ligand complexes and can support future studies including studies o...